Python front-end (pyfabm)
FABM comes with a pyfabm package for the python programming language.
This package enables you to access
FABM’s biogeochemical models directly
from python. For instance, you can enumerate a model’s parameters and variables,
or obtain temporal derivatives and diagnostics for any given environment and
model state. In combination with a python-based time integration scheme
(e.g., scipy.integrate.odeint), this also allows you to perform model
simulations. More information about pyfabm can be found on the
FABM wiki page.
Below you can find brief instructions on how to use pyfabm with ERSEM.
Running pyfabm tutorial
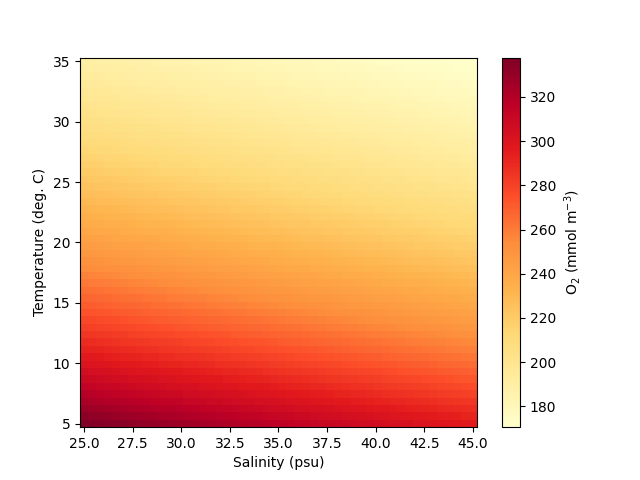
To demonstrate how pyfabm can be used with ERSEM, we will calculate the
oxygen saturation concentration as a function of temperature and salinity
using the method implemented in ERSEM, which is taken from [3].
Note
The following script requires matplotlib to be installed. This can
easily be done via pip in the following way:
python -m pip install matplotlib
With pyfabm installed, this can be achieved using the following code:
1from matplotlib import cm
2import matplotlib.pylab as plt
3import numpy as np
4import os
5import pyfabm
6
7def main():
8 """
9 Run pyfabm-ERSEM tutorial
10
11 :param model_path: Full path to netCDF model output
12 :type model_path: str
13
14 :return: list of oxygen saturation concentrations
15 :rtype: list
16 """
17 dir_path = os.path.dirname(os.path.realpath(__file__))
18 ersem_dir = os.path.dirname(os.path.dirname(dir_path))
19
20 # Path to ERSEM yaml file
21 ersem_yaml_file = os.path.join(ersem_dir,
22 'testcases',
23 'fabm-ersem-15.06-L4-noben-docdyn-iop.yaml')
24
25 if not os.path.isfile(ersem_yaml_file):
26 raise RuntimeError("Could not find Ersem yaml file with the "
27 "{}".format(ersem_yaml_file))
28
29 # Create model
30 model = pyfabm.Model(ersem_yaml_file)
31
32 # Configure the environment
33 model.findDependency('longitude').value = -4.15
34 model.findDependency('latitude').value = 50.25
35 model.findDependency('number_of_days_since_start_of_the_year').value = 0.
36 model.findDependency('temperature').value = 10.
37 model.findDependency('wind_speed').value = 1.
38 model.findDependency('surface_downwelling_shortwave_flux').value = 50.
39 model.findDependency('practical_salinity').value = 35.
40 model.findDependency('pressure').value = 10.
41 model.findDependency('density').value = 1035.
42 model.findDependency('mole_fraction_of_carbon_dioxide_in_air').value = 280.
43 model.findDependency('absorption_of_silt').value = 0.07
44 model.findDependency('bottom_stress').value = 0.
45 model.findDependency('cell_thickness').value = 1.
46 model.setCellThickness(1)
47
48 # Verify the model is ready to be used
49 assert model.checkReady(), 'One or more model dependencies have not been fulfilled.'
50
51 # Define ranges over which temperature and salinity will be varied
52 n_points = 50
53 temperature_array = np.linspace(5, 35, n_points)
54 salinity_array = np.linspace(25, 45, n_points)
55
56 # Create an array in which to store oxygen_saturation_concentrations
57 oxygen_saturation_concentration = np.empty((n_points, n_points), dtype=float)
58
59 # Calculate oxygen saturation concentrations using ERSEM
60 for t_idx, t in enumerate(temperature_array):
61 model.findDependency('temperature').value = t
62 for s_idx, s in enumerate(salinity_array):
63 model.findDependency('practical_salinity').value = s
64 _ = model.getRates()
65 oxygen_saturation_concentration[t_idx, s_idx] = \
66 model.findDiagnosticVariable('O2/osat').value
67
68 # Create the figure
69 figure = plt.figure()
70 axes = plt.gca()
71
72 # Set color map
73 cmap = cm.get_cmap('YlOrRd')
74
75 # Plot
76 plot = axes.pcolormesh(salinity_array,
77 temperature_array,
78 oxygen_saturation_concentration,
79 shading='auto',
80 cmap=cmap)
81
82 axes.set_xlabel('Salinity (psu)')
83 axes.set_ylabel('Temperature (deg. C)')
84
85
86
87 # Add colour bar
88 cbar = figure.colorbar(plot)
89 cbar.set_label('O$_{2}$ (mmol m$^{-3}$)')
90
91 plt.show()
92
93 return oxygen_saturation_concentration
94
95if __name__ == "__main__":
96 main()
In additions, example Jupyter notebooks that use the Python front-end can
be found in <FABMDIR>/testcases/python and more examples can be found
on the FABM wiki.